Resources
In collaboration with international partners we have developed a suite of tools to study gene function in wheat including:
- www.wheat-expression.com – gene expression atlas
- http://bar.utoronto.ca/efp_wheat/cgi-bin/efpWeb.cgi – wheat eFP browser
- www.wheat-training.com – wheat training website
- www.wheat-tilling.com – wheat TILLING mutants
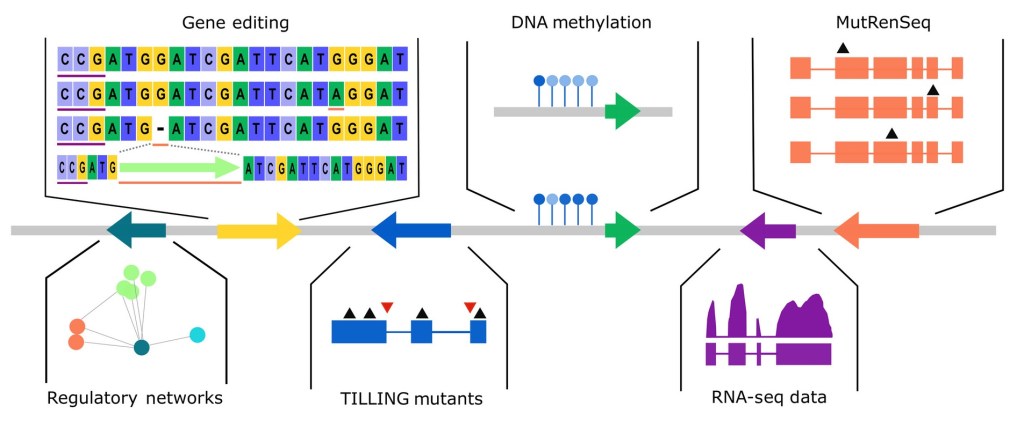
For information on how to use these resources and others related to wheat please visit our training website and read our roadmap for working with wheat.
Code
Code from our pre-prints and publications is available on our Github page
- Study on gene expression during flag leaf senescence (code) (manuscript)
- Study on homoeolog expression in wheat (code) (manuscript)
- Study on TF triad retention (code) (manuscript)
- Study on gene expression during flag leaf senescence in NAM RNAi plants (code) (manuscript)
- Study about NAC5 downstream target genes from DAP-seq (code) (manuscript)
- Study about TaMET1-1 (code) (manuscript)
- Study on homoeolog expression bias in biparental populations (code) (manuscript)
- Study on transcriptional compensation between homoeologs (code) (manuscript)
Protocols
Experimental protocols are described in our publications, with further detailed methods here:
- 3D printer instructions for grain sieve to sort wheat grains according to size: https://www.printables.com/model/890045-grain-sieve
- Protocol to genotype grains and determine parent of origin of alleles: https://dx.doi.org/10.17504/protocols.io.6qpvr84xplmk/v1
- Protocol to identify transcription factor target genes using protoplasts: https://dx.doi.org/10.17504/protocols.io.q26g75bj1lwz/v2
